Single detector, O1 data (restricted prior)#
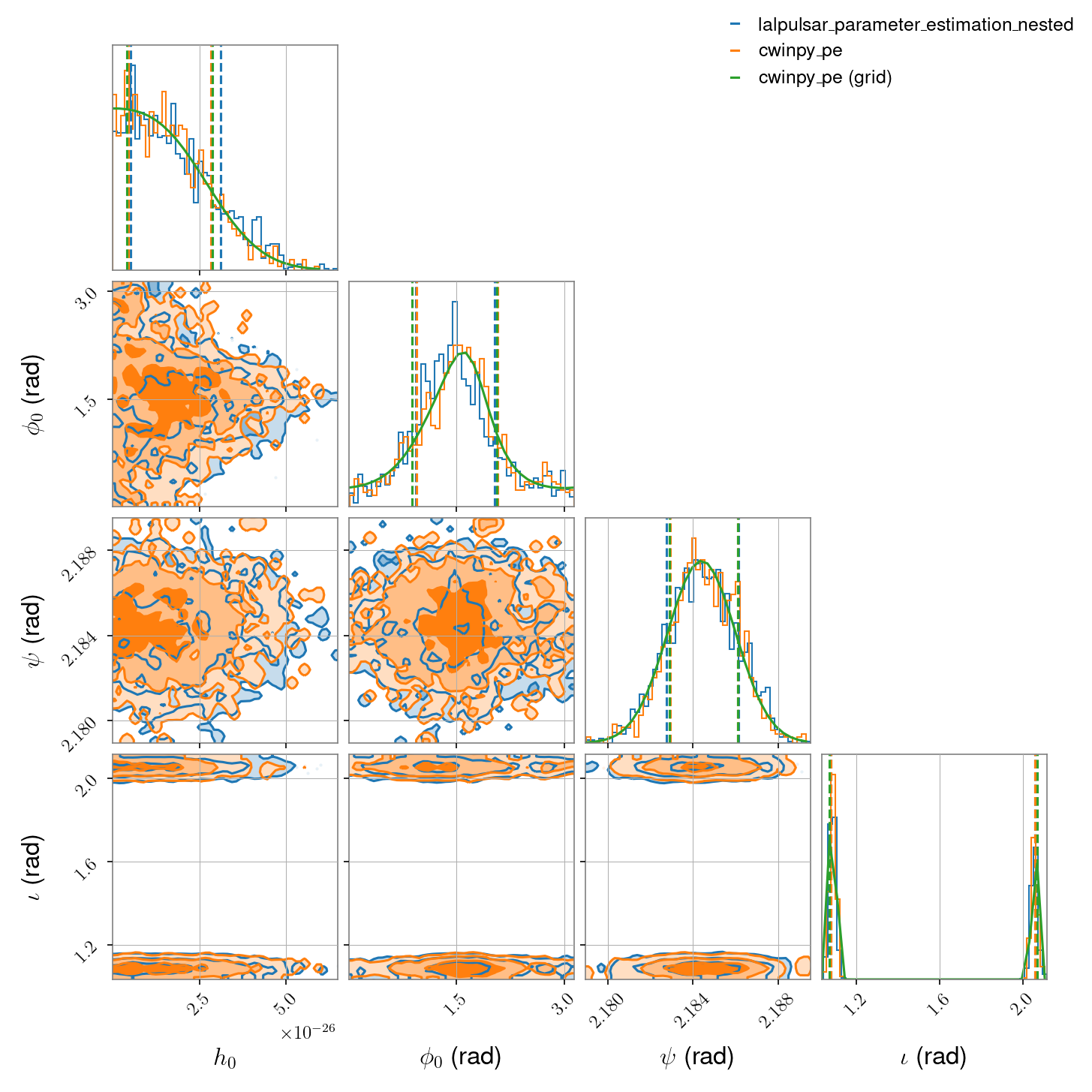
Here we compare lalpulsar_parameter_estimation_nested with cwinpy in the case of real
gravitational-wave data from a single detector (H1 in this case). This data (which can be downloaded
here) comes from the O1 run and has been heterodyned using the parameters for the Crab
pulsar given in this file: J0534+2200.par. The parameters
being estimated are \(h_0\), \(\phi_0\), \(\psi\) and \(\iota\). In this case the
\(h_0\) and \(\phi_0\) parameter have uniform priors, while the \(\psi\) and
\(\iota\) parameters have “restricted” priors. The \(\psi\) prior is a unimodal Gaussian
distribution, while the \(\iota\) prior is a bimodal Gaussian distribution, with values dictated
by fitting the orientation of the Crab’s pulsar wind nebula.
The script for this comparison, using the dynesty nested sampling algorithm, is shown at the bottom of the page. It produces the following comparison data:
Method |
\(\ln{(Z)}\) |
\(\ln{(Z)}\) noise |
\(\ln{}\) Odds |
|---|---|---|---|
|
12126322.815 |
12126328.733 |
-5.918±0.077 |
|
12048289.941 |
12048295.884 |
-5.943±0.137 |
|
12048290.665 |
-5.218 |
Method |
\(h_0\) |
\(\phi_0\) (rad) |
\(\psi\) (rad) |
\(\cos{\iota}\) |
|---|---|---|---|---|
|
1.78±1.24×10-26 |
1.48±0.60 |
2.18±0.00 |
0.07±0.46 |
90% credible intervals |
[0.16, 4.18]×10-26 |
[0.50, 2.69] |
[2.18, 2.19] |
[-0.48, 0.48] |
|
1.66±1.15×10-26 |
1.54±0.61 |
2.18±0.00 |
0.09±0.46 |
90% credible intervals |
[0.14, 3.78]×10-26 |
[0.51, 2.72] |
[2.18, 2.19] |
[-0.48, 0.49] |
Method |
\(h_0\) |
\(\phi_0\) (rad) |
\(\psi\) (rad) |
\(\cos{\iota}\) |
\(\ln{(L)}\) max |
|---|---|---|---|---|---|
|
2.51×10-26 |
1.54 |
2.18 |
1.08 |
12126329.77 |
|
1.95×10-26 |
1.74 |
2.18 |
1.09 |
12048297.03 |
#!/usr/bin/env python
"""
Compare cwinpy with lalpulsar_parameter_estimation_nested for O1 data
from a single detector (H1). This uses data for the Crab pulsar, but restricts
the prior ranges on iota and psi based on observational constraints from the
pulsar wind nebula.
"""
import os
import subprocess as sp
import h5py
import matplotlib
import numpy as np
from astropy.utils.data import download_file
from bilby.core.prior import (
Gaussian,
MultivariateGaussian,
MultivariateGaussianDist,
Uniform,
)
from comparitors import comparisons
from lalinference import LALInferenceHDF5PosteriorSamplesDatasetName
from lalinference.io import read_samples
from matplotlib import pyplot as plt
from cwinpy.pe import pe
from cwinpy.plot import Plot
matplotlib.use("Agg")
# URL for ephemeris files
DOWNLOAD_URL = "https://git.ligo.org/lscsoft/lalsuite/raw/master/lalpulsar/lib/{}"
label = "single_detector_O1_data_restricted_prior"
outdir = "outputs"
if not os.path.isdir(outdir):
os.makedirs(outdir)
# set the par file
parfile = os.path.join("data", "J0534+2200.par")
# set the data file
hetfile = os.path.join("data", "O1_Crab_H1.txt.gz")
# set the detector name
detector = "H1"
# create priors
phi0range = [0.0, np.pi]
psimeansigma = [2.1844, 1.6e-3] # mean and standard deviation of Gaussian prior
h0range = [0.0, 1e-23]
# set prior for lalpulsar_parameter_estimation_nested
priorfile = os.path.join(outdir, "{}_prior.txt".format(label))
priorcontent = """H0 uniform {} {}
PHI0 uniform {} {}
PSI gaussian {} {}
"""
# bi-model Gaussian distribution for ioa
iotamodes = 2 # 2 modes
iotameans = [1.085, 2.0566]
iotavars = [0.00022201, 0.00022201]
iotaweights = [1, 1]
iotaprior = "IOTA gmm {} [[{}],[{}]] [[[{}]],[[{}]]] [{},{}]\n"
with open(priorfile, "w") as fp:
fp.write(priorcontent.format(*(h0range + phi0range + psimeansigma)))
fp.write(iotaprior.format(*([iotamodes] + iotameans + iotavars + iotaweights)))
# set prior for bilby
priors = {}
priors["h0"] = Uniform(h0range[0], h0range[1], "h0", latex_label=r"$h_0$")
priors["phi0"] = Uniform(
phi0range[0], phi0range[1], "phi0", latex_label=r"$\phi_0$", unit="rad"
)
priors["psi"] = Gaussian(
psimeansigma[0], psimeansigma[1], "psi", latex_label=r"$\psi$", unit="rad"
)
iotadist = MultivariateGaussianDist(
names=["iota"],
nmodes=iotamodes,
mus=[[iotameans[0]], [iotameans[1]]],
covs=[[[iotavars[0]]], [[iotavars[1]]]],
weights=iotaweights,
)
priors["iota"] = MultivariateGaussian(
iotadist, "iota", latex_label=r"$\iota$", unit="rad"
)
# run lalpulsar_parameter_estimation_nested
try:
execpath = os.environ["CONDA_PREFIX"]
except KeyError:
raise KeyError(
"Please work in a conda environment with lalsuite and cwinpy installed"
)
execpath = os.path.join(execpath, "bin")
lppen = os.path.join(execpath, "lalpulsar_parameter_estimation_nested")
n2p = os.path.join(execpath, "lalinference_nest2pos")
Nlive = 1000 # number of nested sampling live points
Nmcmcinitial = 0 # set to 0 so that prior samples are not resampled
outfile = os.path.join(outdir, "{}_nest.hdf".format(label))
# set ephemeris files
efile = download_file(DOWNLOAD_URL.format("earth00-40-DE405.dat.gz"), cache=True)
sfile = download_file(DOWNLOAD_URL.format("sun00-40-DE405.dat.gz"), cache=True)
tfile = download_file(DOWNLOAD_URL.format("te405_2000-2040.dat.gz"), cache=True)
# set the command line arguments
runcmd = " ".join(
[
lppen,
"--verbose",
"--input-files",
hetfile,
"--detectors",
detector,
"--par-file",
parfile,
"--prior-file",
priorfile,
"--Nlive",
"{}".format(Nlive),
"--Nmcmcinitial",
"{}".format(Nmcmcinitial),
"--outfile",
outfile,
"--ephem-earth",
efile,
"--ephem-sun",
sfile,
"--ephem-timecorr",
tfile,
]
)
with sp.Popen(
runcmd,
stdout=sp.PIPE,
stderr=sp.PIPE,
shell=True,
bufsize=1,
universal_newlines=True,
) as p:
for line in p.stderr:
print(line, end="")
# convert nested samples to posterior samples
outpost = os.path.join(outdir, "{}_post.hdf".format(label))
runcmd = " ".join([n2p, "-p", outpost, outfile])
with sp.Popen(
runcmd,
stdout=sp.PIPE,
stderr=sp.PIPE,
shell=True,
bufsize=1,
universal_newlines=True,
) as p:
for line in p.stdout:
print(line, end="")
# get posterior samples
post = read_samples(outpost, tablename=LALInferenceHDF5PosteriorSamplesDatasetName)
lp = len(post["H0"])
postsamples = np.zeros((lp, len(priors)))
for i, p in enumerate(priors.keys()):
postsamples[:, i] = post[p.upper()]
# get evidence
hdf = h5py.File(outpost, "r")
a = hdf["lalinference"]["lalinference_nest"]
evsig = a.attrs["log_evidence"]
evnoise = a.attrs["log_noise_evidence"]
hdf.close()
# run bilby via the pe interface
runner = pe(
data_file=hetfile,
par_file=parfile,
prior=priors,
detector=detector,
outdir=outdir,
label=label,
)
result = runner.result
# evaluate the likelihood on a grid
gridpoints = 30
grid_size = dict()
for p in priors.keys():
grid_size[p] = np.linspace(
np.min(result.posterior[p]), np.max(result.posterior[p]), gridpoints
)
grunner = pe(
data_file=hetfile,
par_file=parfile,
prior=priors,
detector=detector,
outdir=outdir,
label=label,
grid=True,
grid_kwargs={"grid_size": grid_size},
)
grid = grunner.grid
# output comparisons
comparisons(label, outdir, grid, priors, cred=0.9)
# create results plot
allresults = {
"lalpulsar_parameter_estimation_nested": outpost,
"cwinpy_pe": result,
"cwinpy_pe (grid)": grid,
}
colors = {
key: plt.rcParams["axes.prop_cycle"].by_key()["color"][i]
for i, key in enumerate(allresults.keys())
}
plot = Plot(
results=allresults,
parameters=list(priors.keys()),
plottype="corner",
)
plot.plot(
bins=50,
smooth=0.9,
quantiles=[0.16, 0.84],
levels=(1 - np.exp(-0.5), 1 - np.exp(-2), 1 - np.exp(-9 / 2.0)),
fill_contours=True,
colors=colors,
)
plot.savefig(os.path.join(outdir, "{}_corner.png".format(label)), dpi=150)