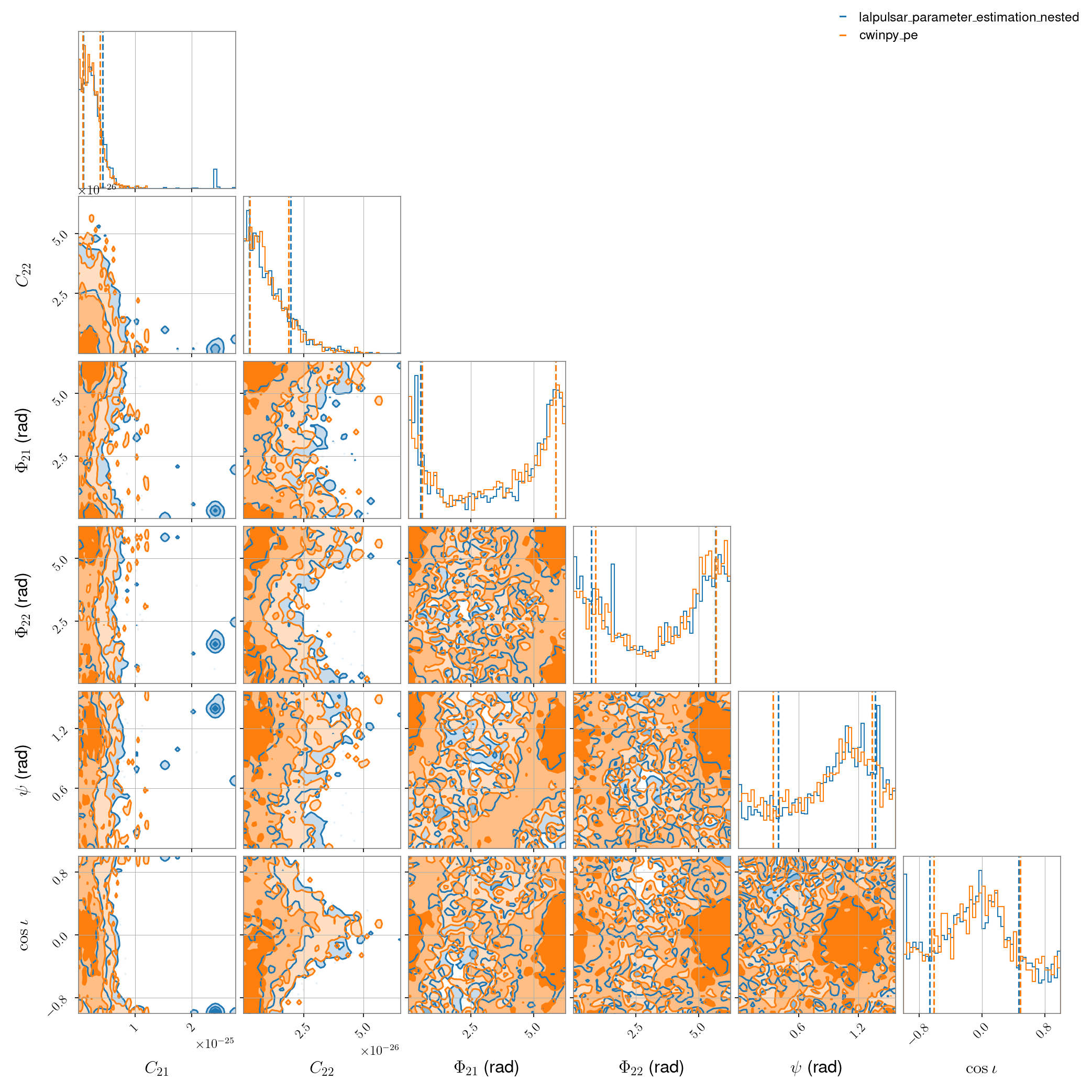
Single detector, noise-only, two harmonics#
Here we compare lalpulsar_parameter_estimation_nested with cwinpy in the case of
simulated Gaussian noise from a single detector (H1 in this case) where the signal is composed of
two harmonics: one at the source rotation frequency and one at twice the rotation frequency. The
parameters being estimated are \(C_{21}\), \(C_{22}\), \(\Phi_{21}\), \(\Phi_{22}\)
\(\psi\) and \(\cos{\iota}\), all with uniform priors.
The script for this comparison, using the dynesty nested sampling algorithm, is shown at the bottom of the page. It produces the following comparison data:
Method |
\(\ln{(Z)}\) |
\(\ln{(Z)}\) noise |
\(\ln{}\) Odds |
|---|---|---|---|
|
323901.783 |
323913.948 |
-12.165±0.112 |
|
323583.166 |
323595.270 |
-12.104±0.192 |
Method |
\(C_{21}\) |
\(C_{22}\) |
\(\Phi_{21}\) (rad) |
\(\Phi_{22}\) (rad) |
\(\psi\) (rad) |
\(\cos{\iota}\) |
|---|---|---|---|---|---|---|
|
3.04±3.82×10-26 |
1.09±0.90×10-26 |
3.62±2.24 |
3.29±2.09 |
0.92±0.42 |
-0.08±0.52 |
90% credible intervals |
[0.28, 6.15]×10-26 |
[0.09, 2.84]×10-26 |
[0.16, 6.15] |
[0.19, 6.06] |
[0.12, 1.49] |
[-0.92, 0.83] |
|
2.35±1.68×10-26 |
1.07±0.88×10-26 |
3.61±2.17 |
3.50±2.06 |
0.89±0.43 |
-0.06±0.51 |
90% credible intervals |
[0.19, 5.25]×10-26 |
[0.09, 2.82]×10-26 |
[0.12, 6.12] |
[0.23, 6.11] |
[0.09, 1.49] |
[-0.88, 0.83] |
Method |
\(C_{21}\) |
\(C_{22}\) |
\(\Phi_{21}\) (rad) |
\(\Phi_{22}\) (rad) |
\(\psi\) (rad) |
\(\cos{\iota}\) |
\(\ln{(L)}\) max |
|---|---|---|---|---|---|---|---|
|
3.29×10-26 |
1.89×10-26 |
6.07 |
5.70 |
1.10 |
0.24 |
323916.62 |
|
2.86×10-26 |
9.66×10-27 |
5.99 |
6.13 |
1.07 |
-0.11 |
323597.98 |
#!/usr/bin/env python
"""
Compare cwinpy with lalpulsar_parameter_estimation_nested for noise-only
data for a single detector with two harmonics.
"""
import os
import subprocess as sp
import h5py
import matplotlib
import numpy as np
from astropy.utils.data import download_file
from bilby.core.prior import Uniform
from comparitors import comparisons_two_harmonics
from lalinference import LALInferenceHDF5PosteriorSamplesDatasetName
from lalinference.io import read_samples
from matplotlib import pyplot as plt
from cwinpy import HeterodynedData
from cwinpy.pe import pe
from cwinpy.plot import Plot
matplotlib.use("Agg")
# URL for ephemeris files
DOWNLOAD_URL = "https://git.ligo.org/lscsoft/lalsuite/raw/master/lalpulsar/lib/{}"
# create a fake pulsar parameter file
parcontent = """\
PSRJ J0123+3456
RAJ 01:23:45.6789
DECJ 34:56:54.321
F0 567.89
F1 -1.2e-12
PEPOCH 56789
C21 0
C22 0
COSIOTA 0
PSI 0
PHI21 0
PHI22 0
"""
label = "single_detector_noise_only_two_harmonics"
outdir = "outputs"
if not os.path.isdir(outdir):
os.makedirs(outdir)
# add content to the par file
parfile = os.path.join(outdir, "{}.par".format(label))
with open(parfile, "w") as fp:
fp.write(parcontent)
# create some fake heterodyned data
detector = "H1" # the detector to use
asds = [1e-24, 2e-24] # noise amplitude spectral densities
times = np.linspace(1000000000.0, 1000086340.0, 1440) # times
harmonics = [1, 2]
hetfiles = []
for harmonic, asd in zip(harmonics, asds):
het = HeterodynedData(
times=times, par=parfile, fakeasd=asd, detector=detector, freqfactor=harmonic
)
# output the data
hetfile = os.path.join(
outdir, "{}_{}_{}_data.txt".format(label, detector, harmonic)
)
het.write(hetfile)
hetfiles.append(hetfile)
# create priors
phi21range = [0.0, 2.0 * np.pi]
phi22range = [0.0, 2.0 * np.pi]
psirange = [0.0, np.pi / 2.0]
cosiotarange = [-1.0, 1.0]
c21range = [0.0, 1e-23]
c22range = [0.0, 1e-23]
# set prior for lalpulsar_parameter_estimation_nested
priorfile = os.path.join(outdir, "{}_prior.txt".format(label))
priorcontent = """C21 uniform {} {}
C22 uniform {} {}
PHI21 uniform {} {}
PHI22 uniform {} {}
PSI uniform {} {}
COSIOTA uniform {} {}
"""
with open(priorfile, "w") as fp:
fp.write(
priorcontent.format(
*(c21range + c22range + phi21range + phi22range + psirange + cosiotarange)
)
)
# set prior for bilby
priors = {}
priors["c21"] = Uniform(c21range[0], c21range[1], "c21", latex_label=r"$C_{21}$")
priors["c22"] = Uniform(c22range[0], c22range[1], "c22", latex_label=r"$C_{22}$")
priors["phi21"] = Uniform(
phi21range[0], phi21range[1], "phi21", latex_label=r"$\Phi_{21}$", unit="rad"
)
priors["phi22"] = Uniform(
phi22range[0], phi22range[1], "phi22", latex_label=r"$\Phi_{22}$", unit="rad"
)
priors["psi"] = Uniform(
psirange[0], psirange[1], "psi", latex_label=r"$\psi$", unit="rad"
)
priors["cosiota"] = Uniform(
cosiotarange[0], cosiotarange[1], "cosiota", latex_label=r"$\cos{\iota}$"
)
# run lalpulsar_parameter_estimation_nested
try:
execpath = os.environ["CONDA_PREFIX"]
except KeyError:
raise KeyError(
"Please work in a conda environment with lalsuite and cwinpy installed"
)
execpath = os.path.join(execpath, "bin")
lppen = os.path.join(execpath, "lalpulsar_parameter_estimation_nested")
n2p = os.path.join(execpath, "lalinference_nest2pos")
Nlive = 1000 # number of nested sampling live points
Nmcmcinitial = 0 # set to 0 so that prior samples are not resampled
outfile = os.path.join(outdir, "{}_nest.hdf".format(label))
# set ephemeris files
efile = download_file(DOWNLOAD_URL.format("earth00-40-DE405.dat.gz"), cache=True)
sfile = download_file(DOWNLOAD_URL.format("sun00-40-DE405.dat.gz"), cache=True)
tfile = download_file(DOWNLOAD_URL.format("te405_2000-2040.dat.gz"), cache=True)
# set the command line arguments
runcmd = " ".join(
[
lppen,
"--verbose",
"--input-files",
",".join(hetfiles),
"--detectors",
detector,
"--par-file",
parfile,
"--prior-file",
priorfile,
"--Nlive",
"{}".format(Nlive),
"--harmonics",
",".join([str(harmonic) for harmonic in harmonics]),
"--Nmcmcinitial",
"{}".format(Nmcmcinitial),
"--outfile",
outfile,
"--ephem-earth",
efile,
"--ephem-sun",
sfile,
"--ephem-timecorr",
tfile,
]
)
p = sp.Popen(runcmd, stdout=sp.PIPE, stderr=sp.PIPE, shell=True)
out, err = p.communicate()
# convert nested samples to posterior samples
outpost = os.path.join(outdir, "{}_post.hdf".format(label))
runcmd = " ".join([n2p, "-p", outpost, outfile])
with sp.Popen(
runcmd,
stdout=sp.PIPE,
stderr=sp.PIPE,
shell=True,
bufsize=1,
universal_newlines=True,
) as p:
for line in p.stdout:
print(line, end="")
# get posterior samples
post = read_samples(outpost, tablename=LALInferenceHDF5PosteriorSamplesDatasetName)
lp = len(post["H0"])
postsamples = np.zeros((lp, len(priors)))
for i, p in enumerate(priors.keys()):
postsamples[:, i] = post[p.upper()]
# get evidence
hdf = h5py.File(outpost, "r")
a = hdf["lalinference"]["lalinference_nest"]
evsig = a.attrs["log_evidence"]
evnoise = a.attrs["log_noise_evidence"]
hdf.close()
# run bilby via the pe interface
runner = pe(
data_file_1f=hetfiles[0],
data_file_2f=hetfiles[1],
par_file=parfile,
prior=priors,
detector=detector,
outdir=outdir,
label=label,
)
result = runner.result
# output comparisons
comparisons_two_harmonics(label, outdir, priors, cred=0.9)
# create results plot
allresults = {
"lalpulsar_parameter_estimation_nested": outpost,
"cwinpy_pe": result,
}
colors = {
key: plt.rcParams["axes.prop_cycle"].by_key()["color"][i]
for i, key in enumerate(allresults.keys())
}
plot = Plot(
results=allresults,
parameters=list(priors.keys()),
plottype="corner",
)
plot.plot(
bins=50,
smooth=0.9,
quantiles=[0.16, 0.84],
levels=(1 - np.exp(-0.5), 1 - np.exp(-2), 1 - np.exp(-9 / 2.0)),
fill_contours=True,
colors=colors,
)
plot.savefig(os.path.join(outdir, "{}_corner.png".format(label)), dpi=150)